-
 Fruit Photosynthesis: More to Know about Where, How and Why
Fruit Photosynthesis: More to Know about Where, How and Why -
 The Growth Oscillator and Plant Stomata: An Open and Shut Case
The Growth Oscillator and Plant Stomata: An Open and Shut Case -
 Tracking Permeation of Dimethyl Sulfoxide (DMSO) in Mentha × piperita Shoot Tips Using Coherent Raman Microscopy
Tracking Permeation of Dimethyl Sulfoxide (DMSO) in Mentha × piperita Shoot Tips Using Coherent Raman Microscopy -
 Genetic Mapping of Seven Kinds of Locus for Resistance to Asian Soybean Rust
Genetic Mapping of Seven Kinds of Locus for Resistance to Asian Soybean Rust -
 Effect of Growth Stages on Anthocyanins and Polyphenols in the Root System of Sweet Potato
Effect of Growth Stages on Anthocyanins and Polyphenols in the Root System of Sweet Potato
Journal Description
Plants
Plants
is an international, scientific, peer-reviewed, open access journal published semimonthly online by MDPI. The Australian Society of Plant Scientists (ASPS), the Spanish Phytopathological Society (SEF), the Spanish Society of Plant Physiology (SEFV), the Spanish Society of Horticultural Sciences (SECH) and the Italian Society of Phytotherapy (S.I.Fit.) are affiliated with Plants and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, PubAg, AGRIS, CAPlus / SciFinder, and other databases.
- Journal Rank: JCR - Q1 (Plant Sciences) / CiteScore - Q1 (Plant Science)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.3 days after submission; acceptance to publication is undertaken in 3.1 days (median values for papers published in this journal in the first half of 2023).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
4.5 (2022);
5-Year Impact Factor:
4.8 (2022)
Latest Articles
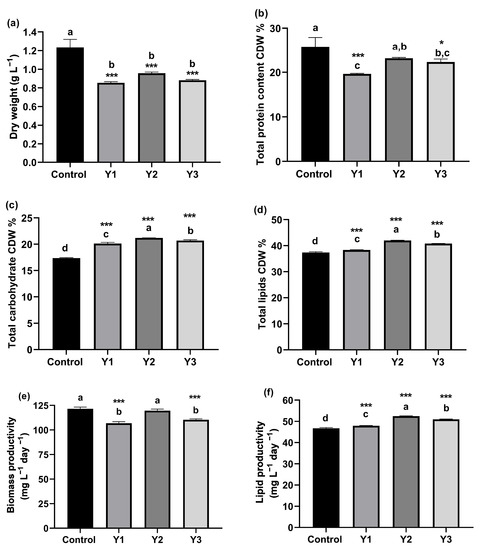
Biodiesel Production from the Marine Alga Nannochloropsis oceanica Grown on Yeast Wastewater and the Effect on Its Biochemical Composition and Gene Expression
Plants 2023, 12(16), 2898; https://doi.org/10.3390/plants12162898 - 08 Aug 2023
Abstract
Microalgae-based biodiesel synthesis is currently not commercially viable due to the high costs of culture realizations and low lipid yields. The main objective of the current study was to determine the possibility of growing Nannochloropsis oceanica on Saccharomyces cerevisiae yeast wastewater for biodiesel
[...] Read more.
Microalgae-based biodiesel synthesis is currently not commercially viable due to the high costs of culture realizations and low lipid yields. The main objective of the current study was to determine the possibility of growing Nannochloropsis oceanica on Saccharomyces cerevisiae yeast wastewater for biodiesel generation at an economical rate. N. oceanica was grown in Guillard F/2 synthetic medium and three dilutions of yeast wastewater (1, 1.25, and 1.5%). Biodiesel properties, in addition to carbohydrate, protein, lipid, dry weight, biomass, lipid productivity, amino acids, and fatty acid methyl ester (FAMEs) content, were analyzed and the quality of the produced biodiesel is assessed. The data revealed the response of N. oceanica to nitrogen-deficiency in the three dilutions of yeast wastewater. N. oceanica in Y2 (1.25%) yeast wastewater dilution exhibited the highest total carbohydrate and lipid percentages (21.19% and 41.97%, respectively), and the highest lipid productivity (52.46 mg L−1 day −1) under nitrogen deficiency in yeast wastewater. The fatty acids profile shows that N. oceanica cultivated in Y2 (1.25%) wastewater dilution provides a significant level of TSFA (47.42%) and can be used as a feedstock for biodiesel synthesis. In addition, N. oceanica responded to nitrogen shortage in wastewater dilutions by upregulating the gene encoding delta-9 fatty acid desaturase (Δ9FAD). As a result, the oleic and palmitoleic acid levels increased in the fatty acid profile of Y2 yeast wastewater dilution, highlighting the increased activity of Δ9FAD enzyme in transforming stearic acid and palmitic acid into oleic acid and palmitoleic acid. This study proved that the Y2 (1.25%) yeast wastewater dilution can be utilized as a growth medium for improving the quantity of specific fatty acids and lipid productivity in N. oceanica that affect biodiesel quality to satisfy global biodiesel requirements.
Full article
(This article belongs to the Special Issue Bioactive Compounds from Marine Plants and Related Sources)
►
Show Figures
Open AccessArticle
Insight into the Molecular Mechanism of Flower Color Regulation in Rhododendron latoucheae Franch: A Multi-Omics Approach
by
, , , , , , and
Plants 2023, 12(16), 2897; https://doi.org/10.3390/plants12162897 - 08 Aug 2023
Abstract
Rhododendron latoucheae Franch. (R. latoucheae) is a valuable woody plant known for its high ornamental value. While purple flowers are a distinct and attractive variant phenotype of R. latoucheae, the underlying mechanism regulating its flower color is still poorly understood.
[...] Read more.
Rhododendron latoucheae Franch. (R. latoucheae) is a valuable woody plant known for its high ornamental value. While purple flowers are a distinct and attractive variant phenotype of R. latoucheae, the underlying mechanism regulating its flower color is still poorly understood. To investigate the molecular regulatory mechanism responsible for the variation in flower color, we selected plants with white-pink and purple petals as the object and conducted analyses of metabolites, key genes, and transcription factors associated with flower color. A combined metabolome–transcriptome analysis was performed, and the expression of key genes was subsequently verified through qRT-PCR experiments. The results of our study demonstrated a significant enrichment of differential metabolites in the flavonoid metabolic pathway. Changes in anthocyanin content followed the same trend as the observed flower color variations, specifically showing significant correlations with the contents of malvidin-3-O-glucoside, dihydromyricetin, gallocatechin, and peonidin-3-O-glucoside. Furthermore, we identified three key structural genes (F3GT1, LAR, ANR) and four transcription factors (bHLH130, bHLH41, bHLH123, MYB4) that are potentially associated with the biosynthesis of flavonoid compounds, thereby influencing the appearance of purple flower color in R. latoucheae.
Full article
(This article belongs to the Special Issue Flower Germplasm Resource and Genetic Breeding)
►▼
Show Figures

Figure 1
Open AccessArticle
The Biochemical Response of Soybean Cultivars Infected by Diaporthe Species Complex
by
, , , , , and
Plants 2023, 12(16), 2896; https://doi.org/10.3390/plants12162896 - 08 Aug 2023
Abstract
Oxidative stress in soybean plants infected with Diaporthe isolates was evaluated in order to select (1) the least aggressive inoculation method, (2) to determine the most aggressive Diaporthe isolate, and (3) to determine the most tolerant soybean cultivar to this isolate. Based on
[...] Read more.
Oxidative stress in soybean plants infected with Diaporthe isolates was evaluated in order to select (1) the least aggressive inoculation method, (2) to determine the most aggressive Diaporthe isolate, and (3) to determine the most tolerant soybean cultivar to this isolate. Based on the present malondialdehyde (MDA) content, the main end product of the lipid peroxidation process, and the biomarker for oxidative stress, the mycelium contact method was chosen as the least aggressive inoculation method, compared to the toothpick method and plug method. The activity of the antioxidant enzymes (superoxide–dismutase (SOD), catalase (CAT), and peroxidase (PX)), the reduced glutathione (GSH) content, and the level of lipid peroxidation (LP) were measured in soybean cv. Sava infected by five different Diaporthe species (DPM1F—D. aspalathi, DPC/KR19—D. caulivora, DPC004NY15—D. eres, 18-DIA-SOY-14—D. gulyae, and PL157A—D. longicolla). The most pathogenic Diaporthe species to cv. Sava was D. eres. The screening of the antioxidant enzymes activity in the leaves of 12 different soybean cultivars (Altona, Atlas, Capital, Chico, CX134, Favorit, Lakota, McCall, Morsoy, Strain, Rubin, and Victoria) infected with D. eres by the mycelium contact inoculation method showed that Capital, McCall, and Morsoy were the cultivars with the highest tolerance to D. eres, followed by Chico, Favorit, Lakota, and Rubin. The most sensitive cultivars were Atlas, CX134, Victoria, and Strain.
Full article
(This article belongs to the Special Issue Fungus and Plant Interactions)
►▼
Show Figures

Figure 1
Open AccessArticle
Physiological Changes and Transcriptomic Analysis throughout On-Tree Fruit Ripening Process in Persimmon (Diospyros kaki L.)
Plants 2023, 12(16), 2895; https://doi.org/10.3390/plants12162895 - 08 Aug 2023
Abstract
The involvement of effectors and transcriptional regulators in persimmon fruit maturation has been mostly approached by the literature under postharvest conditions. In order to elucidate the participation of these genes in the on-tree fruit maturation development, we have collected samples from seven persimmon
[...] Read more.
The involvement of effectors and transcriptional regulators in persimmon fruit maturation has been mostly approached by the literature under postharvest conditions. In order to elucidate the participation of these genes in the on-tree fruit maturation development, we have collected samples from seven persimmon germplasm accessions at different developmental stages until physiological maturation. This study has focused on the expression analysis of 13 genes involved in ethylene biosynthesis and response pathways, as well as the evolution of important agronomical traits such as skin colour, weight, and firmness. Results revealed different gene expression patterns, with genes up- and down-regulated during fruit development progression. A principal component analysis was performed to correlate gene expression with agronomical traits. The decreasing expression of the ethylene biosynthetic genes DkACO1, DkACO2, and DkACS2, in concordance with other sensing (DkERS1) and transduction genes (DkERF18), provides a molecular mechanism for the previously described high production of ethylene in immature detached fruits. On the other side, DkERF8 and DkERF16 are postulated to induce fruit softening and skin colour change during natural persimmon fruit ripening via DkXTH9 and DkPSY activation, respectively. This study provides valuable information for a better understanding of the ethylene signalling pathway and its regulation during on-tree fruit ripening in persimmon.
Full article
(This article belongs to the Special Issue Gene Regulatory Mechanisms of Flower and Fruit Development in Plants)
►▼
Show Figures

Figure 1
Open AccessArticle
Transcriptomic Insight into the Pollen Tube Growth of Olea europaea L. subsp. europaea Reveals Reprogramming and Pollen-Specific Genes Including New Transcription Factors
by
, , , , , and
Plants 2023, 12(16), 2894; https://doi.org/10.3390/plants12162894 - 08 Aug 2023
Abstract
The pollen tube is a key innovation of land plants that is essential for successful fertilisation. Its development and growth have been profusely studied in model organisms, but in spite of the economic impact of olive trees, little is known regarding the genome-wide
[...] Read more.
The pollen tube is a key innovation of land plants that is essential for successful fertilisation. Its development and growth have been profusely studied in model organisms, but in spite of the economic impact of olive trees, little is known regarding the genome-wide events underlying pollen hydration and growth in this species. To fill this gap, triplicate mRNA samples at 0, 1, 3, and 6 h of in vitro germination of olive cultivar Picual pollen were analysed by RNA-seq. A bioinformatics R workflow called RSeqFlow was developed contemplating the best practices described in the literature, covering from expression data filtering to differential expression and clustering, to finally propose hub genes. The resulting olive pollen transcriptome consisted of 22,418 reliable transcripts, where 5364 were differentially expressed, out of which 173 have no orthologue in plants and up to 3 of them might be pollen-specific transcription factors. Functional enrichment revealed a deep transcriptional reprogramming in mature olive pollen that is also dependent on protein stability and turnover to allow pollen tube emergence, with many hub genes related to heat shock proteins and F-box-containing proteins. Reprogramming extends to the first 3 h of growth, including processes consistent with studies performed in other plant species, such as global down-regulation of biosynthetic processes, vesicle/organelle trafficking and cytoskeleton remodelling. In the last stages, growth should be maintained from persistent transcripts. Mature pollen is equipped with transcripts to successfully cope with adverse environments, even though the in vitro growth seems to induce several stress responses. Finally, pollen-specific transcription factors were proposed as probable drivers of pollen germination in olive trees, which also shows an overall increased number of pollen-specific gene isoforms relative to other plants.
Full article
(This article belongs to the Special Issue Applications of Bioinformatics in Plant Resources and Omics)
►▼
Show Figures

Figure 1
Open AccessArticle
Assessment of Soybean Lodging Using UAV Imagery and Machine Learning
Plants 2023, 12(16), 2893; https://doi.org/10.3390/plants12162893 - 08 Aug 2023
Abstract
Plant lodging is one of the most essential phenotypes for soybean breeding programs. Soybean lodging is conventionally evaluated visually by breeders, which is time-consuming and subject to human errors. This study aimed to investigate the potential of unmanned aerial vehicle (UAV)-based imagery and
[...] Read more.
Plant lodging is one of the most essential phenotypes for soybean breeding programs. Soybean lodging is conventionally evaluated visually by breeders, which is time-consuming and subject to human errors. This study aimed to investigate the potential of unmanned aerial vehicle (UAV)-based imagery and machine learning in assessing the lodging conditions of soybean breeding lines. A UAV imaging system equipped with an RGB (red-green-blue) camera was used to collect the imagery data of 1266 four-row plots in a soybean breeding field at the reproductive stage. Soybean lodging scores were visually assessed by experienced breeders, and the scores were grouped into four classes, i.e., non-lodging, moderate lodging, high lodging, and severe lodging. UAV images were stitched to build orthomosaics, and soybean plots were segmented using a grid method. Twelve image features were extracted from the collected images to assess the lodging scores of each breeding line. Four models, i.e., extreme gradient boosting (XGBoost), random forest (RF), K-nearest neighbor (KNN) and artificial neural network (ANN), were evaluated to classify soybean lodging classes. Five data preprocessing methods were used to treat the imbalanced dataset to improve classification accuracy. Results indicate that the preprocessing method SMOTE-ENN consistently performs well for all four (XGBoost, RF, KNN, and ANN) classifiers, achieving the highest overall accuracy (OA), lowest misclassification, higher F1-score, and higher Kappa coefficient. This suggests that Synthetic Minority Oversampling-Edited Nearest Neighbor (SMOTE-ENN) may be a good preprocessing method for using unbalanced datasets and the classification task. Furthermore, an overall accuracy of 96% was obtained using the SMOTE-ENN dataset and ANN classifier. The study indicated that an imagery-based classification model could be implemented in a breeding program to differentiate soybean lodging phenotype and classify lodging scores effectively.
Full article
(This article belongs to the Special Issue Advances in Sensor Systems and Data Analysis for Crop Phenotyping)
►▼
Show Figures

Figure 1
Open AccessArticle
Irrigation Strategies with Controlled Water Deficit in Two Production Cycles of Cotton
by
, , , , , , , , , and
Plants 2023, 12(16), 2892; https://doi.org/10.3390/plants12162892 - 08 Aug 2023
Abstract
Water scarcity is one of the main abiotic factors that limit agricultural production. In this sense, the identification of genotypes tolerant to water deficit associated with irrigation management strategies is extremely important. In this context, the objective of this study was to evaluate
[...] Read more.
Water scarcity is one of the main abiotic factors that limit agricultural production. In this sense, the identification of genotypes tolerant to water deficit associated with irrigation management strategies is extremely important. In this context, the objective of this study was to evaluate the morphology, production, water consumption, and water use efficiency of colored fiber cotton genotypes submitted to irrigation strategies with a water deficit in the phenological phases. Two experiments were conducted in succession. In the first experiment, a randomized block design was used in a 3 × 7 factorial scheme, corresponding to three colored cotton genotypes (BRS Rubi, BRS Jade, and BRS Safira) in seven irrigation management strategies with 40% of the real evapotranspiration (ETr) varying the phenological stages. In the second experiment, the same design was used in a 3 × 10 factorial arrangement (genotypes × irrigation management strategies). The water deficit in the vegetative phase can be used in the first year of cotton cultivation. Among the genotypes, ‘BRS Jade’ is the most tolerant to water deficit in terms of phytomass accumulation and fiber production.
Full article
(This article belongs to the Special Issue Crop Plants Response to Abiotic Stresses)
►▼
Show Figures
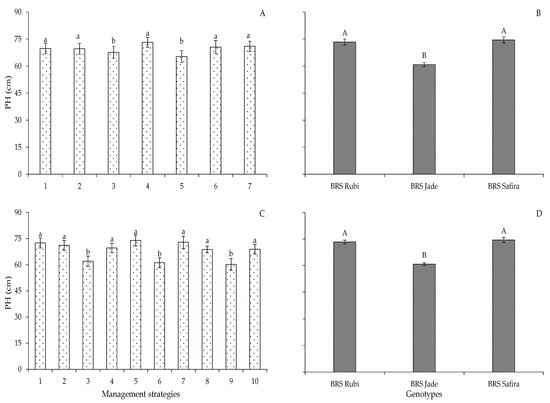
Figure 1
Open AccessReview
Affecting Factors of Plant Phyllosphere Microbial Community and Their Responses to Climatic Warming—A Review
Plants 2023, 12(16), 2891; https://doi.org/10.3390/plants12162891 - 08 Aug 2023
Abstract
Phyllosphere microorganisms are not only an important part of plants, but also an important part of microorganisms. In this review, the function of phyllosphere microorganisms, the assembly mechanism of phyllosphere microorganisms, the driving factors of phyllosphere microbial community structure, and the effects of
[...] Read more.
Phyllosphere microorganisms are not only an important part of plants, but also an important part of microorganisms. In this review, the function of phyllosphere microorganisms, the assembly mechanism of phyllosphere microorganisms, the driving factors of phyllosphere microbial community structure, and the effects of climate warming on phyllosphere microbial community structure were reviewed. Generally, phyllosphere microorganisms have a variety of functions (e.g., fixing nitrogen, promoting plant growth). Although selection and dispersal processes together regulate the assembly of phyllospheric microbial communities, which one of the ecological processes is dominant and how external disturbances alter the relative contributions of each ecological process remains controversial. Abiotic factors (e.g., climatic conditions, geographical location and physical and chemical properties of soil) and biological factors (e.g., phyllosphere morphological structure, physiological and biochemical characteristics, and plant species and varieties) can affect phyllosphere microbial community structure. However, the predominant factors affecting phyllosphere microbial community structure are controversial. Moreover, how climate warming affects the phyllosphere microbial community structure and its driving mechanism have not been fully resolved, and further relevant studies are needed.
Full article
(This article belongs to the Special Issue Microbial Formulations to Increase Crop Production, from Laboratory to Fields)
►▼
Show Figures
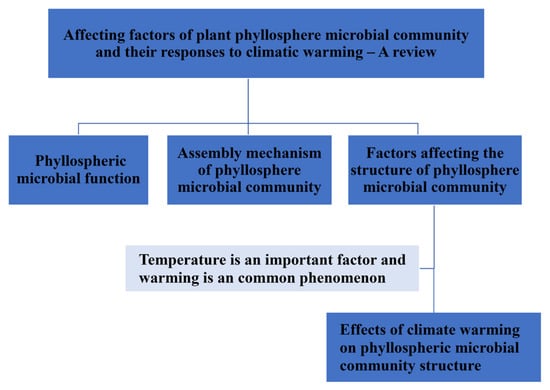
Figure 1
Open AccessArticle
Extraction of Polyphenols from Slovenian Hop (Humulus lupulus L.) Aurora Variety Using Deep Eutectic Solvents: Choice of the Extraction Method vs. Structure of the Solvent
by
, , , , , and
Plants 2023, 12(16), 2890; https://doi.org/10.3390/plants12162890 - 08 Aug 2023
Abstract
Polyphenols from Slovenian hops (Humulus lupulus L.) of the Aurora variety were extracted by different methods and using classical solvents and several deep eutectic solvents (DES) based on choline chloride as the hydrogen bond acceptor component. The obtained extract solutions were analyzed
[...] Read more.
Polyphenols from Slovenian hops (Humulus lupulus L.) of the Aurora variety were extracted by different methods and using classical solvents and several deep eutectic solvents (DES) based on choline chloride as the hydrogen bond acceptor component. The obtained extract solutions were analyzed by HPLC for the content of extracted α- and β-acids and extracted xanthohumol. It was found that choline chloride:phenol DES concentrated aqueous solution had an extraction efficiency close to that of diethyl ether, which is considered one of the best classical extraction solvents for polyphenols from hops. The comparison of the extraction efficiency with other choline chloride-based DESs showed that the chemical similarity of the phenol ring in the solvent DES with the polyphenols in hops may be crucial for a highly efficient extraction with choline chloride:phenol DES. On the other hand, the choice of extraction method and the viscosity of the solvents tested seem to play only a minor role in this respect. As far as we know, this is the first study to attempt to relate extraction efficiency in the extraction of hydrophobic solutes to the compressibility of the DES extractants, the latter of which may be correlated with the extent of hydrophobic hydration around the DES components. In addition, using the heating and stirring method for the preparation of choline chloride-based DES concentrated aqueous solutions we found no support for the occurrence of water in two different roles (in the structural and in the dilution role) in these solvents.
Full article
(This article belongs to the Special Issue Extraction Technologies, Isolation, Separation, and Application of Phytochemicals Volume II)
►▼
Show Figures

Figure 1
Open AccessArticle
Transcriptome Analysis Reveals Changes in Whole Gene Expression, Biological Process, and Molecular Functions Induced by Nickel in Jack Pine (Pinus banksiana)
Plants 2023, 12(15), 2889; https://doi.org/10.3390/plants12152889 - 07 Aug 2023
Abstract
Understanding the genetic response of plants to nickel stress is a necessary step to improving the utility of plants in environmental remediation and restoration. The main objective of this study was to generate whole genome expression profiles of P. banksiana exposed to nickel
[...] Read more.
Understanding the genetic response of plants to nickel stress is a necessary step to improving the utility of plants in environmental remediation and restoration. The main objective of this study was to generate whole genome expression profiles of P. banksiana exposed to nickel ion toxicity compared to reference genotypes. Pinus banksiana seedlings were screened in a growth chamber setting using a high concentration of 1600 mg of nickel per 1 kg of soil. RNA was extracted and sequenced using the Illumina platform, followed by de novo transcriptome assembly. Overall, 25,552 transcripts were assigned gene ontology. The biological processes in water-treated samples were analyzed, and 55% of transcripts were distributed among five categories: DNA metabolic process (19.3%), response to stress (13.3%), response to chemical stimuli (8.7%), signal transduction (7.7%) and response to biotic stimulus (6.0%). For molecular function, the highest percentages of genes were involved in nucleotide binding (27.6%), nuclease activity (27.3%) and kinase activity (10.3%). Sixty-two percent of genes were associated with cellular compartments. Of these genes, 21.7% were found in the plasma membrane, 16.1% in the cytosol, 12.4% with the chloroplast and 11.9% in the extracellular region. Nickel ions induced changes in gene expression, resulting in the emergence of differentially regulated categories. Overall, there were significant changes in gene expression with a total 4128 genes upregulated and 3754 downregulated genes detected in nickel-treated genotypes compared to water-treated control plants. For biological processes, the highest percentage of upregulated genes in plants exposed to nickel were associated with the response to stress (15%), the response to chemicals (11,1%), carbohydrate metabolic processes (7.4%) and catabolic processes (7.4%). The largest proportions of downregulated genes were associated with the biosynthetic process (21%), carbohydrate metabolic process (14.3%), response to biotic stimulus (10.7%) and response to stress (10.7%). For molecular function, genes encoding for enzyme regulatory and hydrolase activities represented the highest proportion (61%) of upregulated gene. The majority of downregulated genes were involved in the biosynthetic processes. Overall, 58% of upregulated genes were located in the extracellular region and the nucleus, while 42% of downregulated genes were localized to the plasma membrane and 33% to the extracellular region. This study represents the first report of a transcriptome from a conifer species treated with nickel.
Full article
(This article belongs to the Special Issue Recent Advances in Plant Genomics and Transcriptome Analysis)
►▼
Show Figures
Figure 1
Open AccessArticle
Chemical Composition, Larvicidal and Molluscicidal Activity of Essential Oils of Six Guava Cultivars Grown in Vietnam
by
, , , , , and
Plants 2023, 12(15), 2888; https://doi.org/10.3390/plants12152888 - 07 Aug 2023
Abstract
Diseases transmitted by mosquitoes and snails cause a large burden of disease in less developed countries, especially those with low-income levels. An approach to control vectors and intermediate hosts based on readily available essential oils, which are friendly to the environment and human
[...] Read more.
Diseases transmitted by mosquitoes and snails cause a large burden of disease in less developed countries, especially those with low-income levels. An approach to control vectors and intermediate hosts based on readily available essential oils, which are friendly to the environment and human health, may be an effective solution for disease control. Guava is a fruit tree grown on a large scale in many countries in the tropics, an area heavily affected by tropical diseases transmitted by mosquitoes and snails. Previous studies have reported that the extracted essential oils of guava cultivars have high yields, possess different chemotypes, and exhibit toxicity to different insect species. Therefore, this study was carried out with the aim of studying the chemical composition and pesticide activities of six cultivars of guava grown on a large scale in Vietnam. The essential oils were extracted by hydrodistillation using a Clevenger-type apparatus for 6 h. The components of the essential oils were determined using gas-chromatography–mass-spectrometry (GC-MS) analysis. Test methods for pesticide activities were performed in accordance with WHO guidelines and modifications. Essential oil samples from Vietnam fell into two composition-based clusters, one of (E)-β-caryophyllene and the other of limonene/(E)-β-caryophyllene. The essential oils PG03 and PG05 show promise as environmentally friendly pesticides when used to control Aedes mosquito larvae with values of 24 h LC50-aegypti of 0.96 and 0.40 µg/mL while 24 h LC50-albopictus of 0.50 and 0.42 µg/mL. These two essential oils showed selective toxicity against Aedes mosquito larvae and were safe against the non-target organism Anisops bouvieri. Other essential oils may be considered as molluscicides against Physa acuta (48 h LC50 of 4.10 to 5.00 µg/mL) and Indoplanorbis exustus (48 h LC50 of 3.85 to 7.71 µg/mL) and with less toxicity to A. bouvieri.
Full article
(This article belongs to the Special Issue Pharmacology and Toxicology of Plants and Their Constituents)
►▼
Show Figures
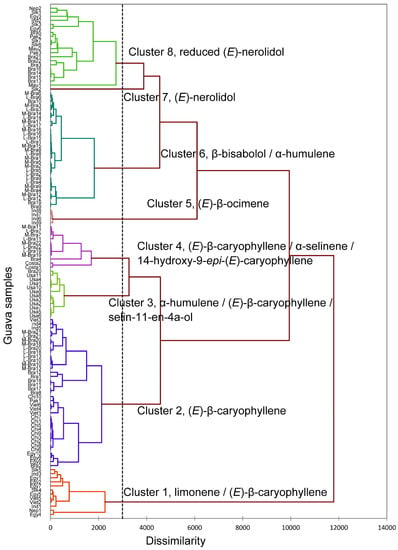
Figure 1
Open AccessArticle
14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction
by
, , , , and
Plants 2023, 12(15), 2887; https://doi.org/10.3390/plants12152887 - 07 Aug 2023
Abstract
The geomagnetic field (GMF) is a natural component of the biosphere, and, during evolution, all organisms experienced its presence while some evolved the ability to perceive magnetic fields (MF). We studied the response of 14-3-3 proteins and the plasma membrane (PM) proton pump
[...] Read more.
The geomagnetic field (GMF) is a natural component of the biosphere, and, during evolution, all organisms experienced its presence while some evolved the ability to perceive magnetic fields (MF). We studied the response of 14-3-3 proteins and the plasma membrane (PM) proton pump H+-ATPase to reduced GMF values by lowering the GMF intensity to a near-null magnetic field (NNMF). Seedling morphology, H+-ATPase activity and content, 14-3-3 protein content, binding to PM and phosphorylation, gene expression, and ROS quantification were assessed in maize (Zea mays) dark-grown seedlings. Phytohormone and melatonin quantification were also assessed by LG-MS/MS. Our results suggest that the GMF regulates the PM H+-ATPase, and that NNMF conditions alter the proton pump activity by reducing the binding of 14-3-3 proteins. This effect was associated with both a reduction in H2O2 and downregulation of genes coding for enzymes involved in ROS production and scavenging, as well as calcium homeostasis. These early events were followed by the downregulation of IAA synthesis and gene expression and the increase in both cytokinin and ABA, which were associated with a reduction in root growth. The expression of the homolog of the MagR gene, ZmISCA2, paralleled that of CRY1, suggesting a possible role of ISCA in maize magnetic induction. Interestingly, melatonin, a widespread molecule present in many kingdoms, was increased by the GMF reduction, suggesting a still unknown role of this molecule in magnetoreception.
Full article
(This article belongs to the Special Issue Plant Responses to Environmental Stresses)
►▼
Show Figures
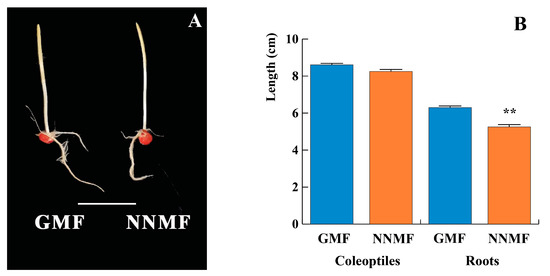
Figure 1
Open AccessArticle
Bottom-Up Effects of Drought-Stressed Cotton Plants on Performance and Feeding Behavior of Aphis gossypii
Plants 2023, 12(15), 2886; https://doi.org/10.3390/plants12152886 - 07 Aug 2023
Abstract
Drought, a major stress for crop plants, is expected to increase in frequency due to climate change. Drought can alter crop growth and levels of secondary plant metabolites, which in turn can affect herbivores, but this latter point is still controversial. This study
[...] Read more.
Drought, a major stress for crop plants, is expected to increase in frequency due to climate change. Drought can alter crop growth and levels of secondary plant metabolites, which in turn can affect herbivores, but this latter point is still controversial. This study used three different polyethylene glycol (PEG-6000) levels (0%, 1%, and 3%) to simulate drought stress and evaluated their effects on cotton plants and the impacts on the performance of the cotton aphid Aphis gossypii. Cotton plants under drought stress showed decreased water content, above-ground biomass, and nitrogen content and increased soluble protein, soluble sugar, and tannin contents. Based on analysis of the developmental time and fecundity data from individuals and at the population level, a significantly lower fecundity and population abundance of A. gossypii were detected on cotton plants with drought stress, which supports the “plant vigor hypothesis”. The poor development of A. gossypii is possibly related to lower xylem sap and phloem ingestion under drought stress. In addition, the increased tannin content of cotton plants induced by drought and lower detoxification enzyme activities of A. gossypii may have affected the responses of aphids to drought-stressed plants. Overall, the results showed that drought stress altered the physiological characteristics of the cotton plants, resulting in adverse bottom-up effects on cotton aphid performances. This implies that the adoption of drip irrigation under plastic film that can help alleviate drought stress may favor the population growth of cotton aphids.
Full article
(This article belongs to the Special Issue Embracing Systems Thinking in Crop Protection Science)
►▼
Show Figures
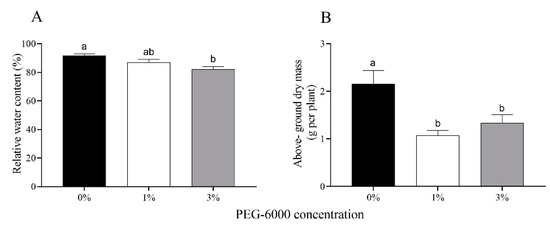
Figure 1
Open AccessArticle
Multiple-Genome-Based Simple Sequence Repeat Is an Efficient and Successful Method in Genotyping and Classifying Different Jujube Germplasm Resources
by
, , , , , , and
Plants 2023, 12(15), 2885; https://doi.org/10.3390/plants12152885 - 07 Aug 2023
Abstract
Jujube (Ziziphus jujuba Mill.) is a commercially important tree native to China, known for its high nutritional value and widespread distribution, as well as its diverse germplasm resources. Being resilient to harsh climatic conditions, the cultivation of jujube could provide a solution
[...] Read more.
Jujube (Ziziphus jujuba Mill.) is a commercially important tree native to China, known for its high nutritional value and widespread distribution, as well as its diverse germplasm resources. Being resilient to harsh climatic conditions, the cultivation of jujube could provide a solution to food insecurity and income for people of arid and semi-arid regions in and outside of China. The evaluation of germplasm resources and genetic diversity in jujube necessitates the use of Simple Sequence Repeat (SSR) markers. SSR markers are highly polymorphic and can be used to evaluate the genetic diversity within and between cultivars of Chinese jujube, and are important for conservation biology, breeding programs, and the discovery of important traits for Chinese jujube improvement in China and abroad. However, traditional methods of SSR development are time-consuming and inadequate to meet the growing research demands. To address this issue, we developed a novel approach called Multiple-Genome-Based SSR identification (MGB-SSR), which utilizes the genomes of three jujube cultivars to rapidly screen for polymorphic SSRs in the jujube genome. Through the screening process, we identified 12 pairs of SSR primers, which were then used to successfully classify 249 jujube genotypes. Based on the genotyping results, a digital ID card was established, enabling the complete identification of all 249 jujube plants. The MGB-SSR approach proved efficient in rapidly detecting polymorphic SSRs within the jujube genome. Notably, this study represents the first successful differentiation of jujube germplasm resources using 12 SSR markers, with 4 markers successfully identifying triploid jujube genotypes. These findings offer valuable information for the classification of Chinese jujube germplasm, thereby providing significant assistance to jujube researchers and breeders in identifying unknown jujube germplasm.
Full article
(This article belongs to the Special Issue Advances in Jujube Research)
►▼
Show Figures
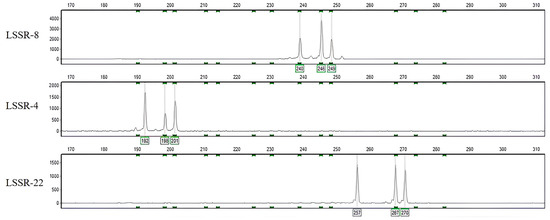
Figure 1
Open AccessArticle
Involvement of NO in V-ATPase Regulation in Cucumber Roots under Control and Cadmium Stress Conditions
Plants 2023, 12(15), 2884; https://doi.org/10.3390/plants12152884 - 07 Aug 2023
Abstract
Nitric oxide (NO) is a signaling molecule that participates in plant adaptation to adverse environmental factors. This study aimed to clarify the role of NO in the regulation of vacuolar H+-ATPase (V-ATPase) in the roots of cucumber seedlings grown under control
[...] Read more.
Nitric oxide (NO) is a signaling molecule that participates in plant adaptation to adverse environmental factors. This study aimed to clarify the role of NO in the regulation of vacuolar H+-ATPase (V-ATPase) in the roots of cucumber seedlings grown under control and Cd stress conditions. In addition, the relationship between NO and salicylic acid (SA), as well as their interrelations with hydrogen sulfide (H2S) and hydrogen peroxide (H2O2), have been verified. The effect of NO on V-ATPase was studied by analyzing two enzyme activities, the expression level of selected VHA genes and the protein level of selected VHA subunits in plants treated with a NO donor (sodium nitroprusside, SNP) and NO biosynthesis inhibitors (tungstate, WO42− and N-nitro-L-arginine methyl ester, L-NAME). Our results indicate that NO functions as a positive regulator of V-ATPase and that this regulation depends on NO generated by nitrate reductase and NOS-like activity. It was found that the mechanism of NO action is not related to changes in the gene expression or protein level of the V-ATPase subunits. The results suggest that in cucumber roots, NO signaling interacts with the SA pathway and, to a lesser extent, with two other known V-ATPase regulators, H2O2 and H2S.
Full article
(This article belongs to the Special Issue Redox Biology in Plants)
►▼
Show Figures
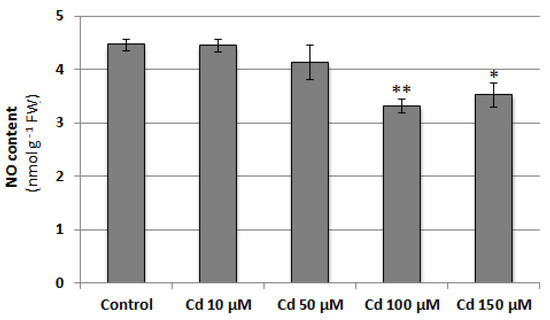
Figure 1
Open AccessArticle
YOLOv7-Plum: Advancing Plum Fruit Detection in Natural Environments with Deep Learning
Plants 2023, 12(15), 2883; https://doi.org/10.3390/plants12152883 - 07 Aug 2023
Abstract
The plum is a kind of delicious and common fruit with high edible value and nutritional value. The accurate and effective detection of plum fruit is the key to fruit number counting and pest and disease early warning. However, the actual plum orchard
[...] Read more.
The plum is a kind of delicious and common fruit with high edible value and nutritional value. The accurate and effective detection of plum fruit is the key to fruit number counting and pest and disease early warning. However, the actual plum orchard environment is complex, and the detection of plum fruits has many problems, such as leaf shading and fruit overlapping. The traditional method of manually estimating the number of fruits and the presence of pests and diseases used in the plum growing industry has disadvantages, such as low efficiency, a high cost, and low accuracy. To detect plum fruits quickly and accurately in a complex orchard environment, this paper proposes an efficient plum fruit detection model based on an improved You Only Look Once version 7(YOLOv7). First, different devices were used to capture high-resolution images of plum fruits growing under natural conditions in a plum orchard in Gulin County, Sichuan Province, and a dataset for plum fruit detection was formed after the manual screening, data enhancement, and annotation. Based on the dataset, this paper chose YOLOv7 as the base model, introduced the Convolutional Block Attention Module (CBAM) attention mechanism in YOLOv7, used Cross Stage Partial Spatial Pyramid Pooling–Fast (CSPSPPF) instead of Cross Stage Partial Spatial Pyramid Pooling(CSPSPP) in the network, and used bilinear interpolation to replace the nearest neighbor interpolation in the original network upsampling module to form the improved target detection algorithm YOLOv7-plum. The tested YOLOv7-plum model achieved an average precision (AP) value of 94.91%, which was a 2.03% improvement compared to the YOLOv7 model. In order to verify the effectiveness of the YOLOv7-plum algorithm, this paper evaluated the performance of the algorithm through ablation experiments, statistical analysis, etc. The experimental results showed that the method proposed in this study could better achieve plum fruit detection in complex backgrounds, which helped to promote the development of intelligent cultivation in the plum industry.
Full article
(This article belongs to the Collection Application of AI in Plants)
►▼
Show Figures
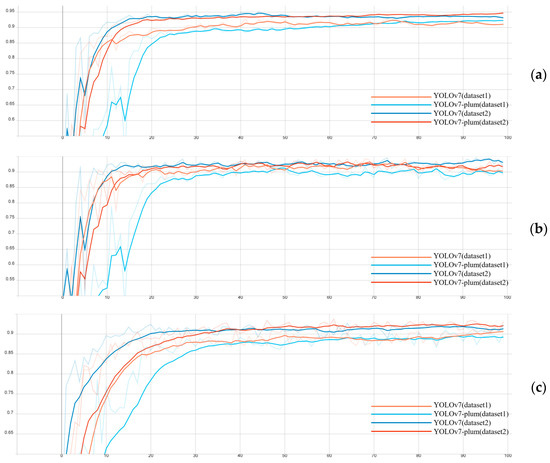
Figure 1
Open AccessArticle
Effect of Soil Acidification on the Production of Se-Rich Tea
by
, , , , , , , and
Plants 2023, 12(15), 2882; https://doi.org/10.3390/plants12152882 - 07 Aug 2023
Abstract
Selenium (Se)-enriched tea is a well-regarded natural beverage that is often consumed for its Se supplementation benefits. However, the production of this tea, particularly in Se-abundant tea plantations, is challenging due to soil acidification. Therefore, this study aimed to investigate the effects of
[...] Read more.
Selenium (Se)-enriched tea is a well-regarded natural beverage that is often consumed for its Se supplementation benefits. However, the production of this tea, particularly in Se-abundant tea plantations, is challenging due to soil acidification. Therefore, this study aimed to investigate the effects of changes in Se under acidified soil conditions. Eight tea plantation soil monitoring sites in Southern Jiangsu were first selected. Simulated acid rain experiments and experiments with different acidification methods were designed and soil pH, as well as various Al-ion and Se-ion concentrations were systematically determined. The data were analyzed using R statistical software, and a correlation analysis was carried out. The results indicated that as the pH value dropped, exchangeable selenium (Exc-Se) and residual selenium (Res-Se) were transformed into acid-soluble selenium (Fmo-Se) and manganese oxide selenium (Om-Se). As the pH increased, exchange state aluminum (Alex) and water-soluble aluminum (Alw) decreased, Fmo-Se and Om-Se declined, and Exc-Se and Res-Se increased, a phenomenon attributed to the weakened substitution of Se ions by Al ions. In the simulated acid rain experiment, P1 compared to the control (CK), the pH value of the YJW tea plantation decreased by 0.13, Exc-Se decreased by 4 ug mg−1, Res-Se decreased by 54.65 ug kg−1, Fmo-Se increased by 2.78 ug mg−1, and Om-Se increased by 5.94 ug mg−1 while Alex increased by 28.53 mg kg−1. The decrease in pH led to an increase in the content of Alex and Alw, which further resulted in the conversion of Exc-Se to Fmo-Se and Om-Se. In various acidification experiments, compared with CK, the pH value of T6 decreased by 0.23, Exc-Se content decreased by 8.35 ug kg−1, Res-Se content decreased by 40.62 ug kg−1, and Fmo-Se content increased by 15.52 ug kg−1 while Alex increased by 33.67 mg kg−1, Alw increased by 1.7 mg kg−1, and Alh decreased by 573.89 mg kg−1. Acidification can trigger the conversion of Exc-Se to Fmo-Se and Om-Se, while the content of available Se may decrease due to the complexation interplay between Alex and Exc-Se. This study provides a theoretical basis for solving the problem of Se-enriched in tea caused by soil acidification.
Full article
(This article belongs to the Special Issue Tea Plants Cultivation)
►▼
Show Figures
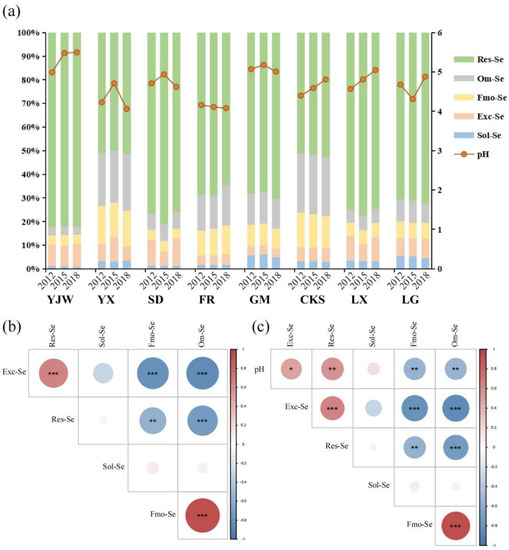
Figure 1
Open AccessArticle
Saline–Alkaline Stress Resistance of Cabernet Sauvignon Grapes Grafted on Different Rootstocks and Rootstock Combinations
Plants 2023, 12(15), 2881; https://doi.org/10.3390/plants12152881 - 06 Aug 2023
Abstract
Grafting the wine grape variety Cabernet Sauvignon onto salinity-tolerant rootstocks can improve salinity tolerance and grape yields in regions with high salinity soils. In this experiment, the effects of different rootstocks and rootstock combinations on the saline–alkaline stress (modified Hoagland nutrient solution +
[...] Read more.
Grafting the wine grape variety Cabernet Sauvignon onto salinity-tolerant rootstocks can improve salinity tolerance and grape yields in regions with high salinity soils. In this experiment, the effects of different rootstocks and rootstock combinations on the saline–alkaline stress (modified Hoagland nutrient solution + 50 mmol L−1 (NaCl + NaHCO3)) of Cabernet Sauvignon were studied. Correlation and principal component analyses were conducted on several physiological indicators of saline–alkaline stress. Salinity limited biomass accumulation, induced damage to the plant membrane, reduced the chlorophyll content and photosynthetic capacity of plants, and increased the content of malondialdehyde, sodium (Na+)/potassium (K+) ratio, and antioxidant enzyme activities (superoxide dismutase, peroxidase, and catalase). Significant differences in several indicators were observed among the experimental groups. The results indicate that the saline–alkaline tolerance of Cabernet Sauvignon after grafting was the same as that of the rootstock, indicating that the increased resistance of Cabernet Sauvignon grapes to saline–alkaline stress stems from the transferability of the saline–alkaline stress resistance of the rootstock to the scion.
Full article
(This article belongs to the Section Plant Response to Abiotic Stress and Climate Change)
►▼
Show Figures
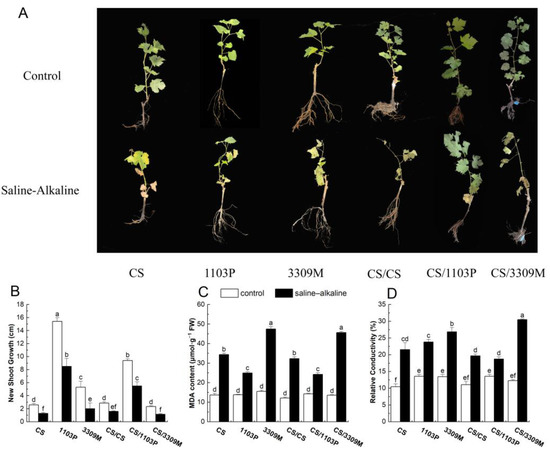
Figure 1
Open AccessArticle
PlantMetSuite: A User-Friendly Web-Based Tool for Metabolomics Analysis and Visualisation
Plants 2023, 12(15), 2880; https://doi.org/10.3390/plants12152880 - 06 Aug 2023
Abstract
The advancement of mass spectrometry technologies has revolutionised plant metabolomics research by enabling the acquisition of raw metabolomics data. However, the identification, analysis, and visualisation of these data require specialised tools. Existing solutions lack a dedicated plant-specific metabolite database and pose usability challenges.
[...] Read more.
The advancement of mass spectrometry technologies has revolutionised plant metabolomics research by enabling the acquisition of raw metabolomics data. However, the identification, analysis, and visualisation of these data require specialised tools. Existing solutions lack a dedicated plant-specific metabolite database and pose usability challenges. To address these limitations, we developed PlantMetSuite, a web-based tool for comprehensive metabolomics analysis and visualisation. PlantMetSuite encompasses interactive bioinformatics tools and databases specifically tailored to plant metabolomics data, facilitating upstream-to-downstream analysis in metabolomics and supporting integrative multi-omics investigations. PlantMetSuite can be accessed directly through a user’s browser without the need for installation or programming skills. The tool is freely available and will undergo regular updates and expansions to incorporate additional libraries and newly published metabolomics analysis methods. The tool’s significance lies in empowering researchers with an accessible and customisable platform for unlocking plant metabolomics insights.
Full article
(This article belongs to the Special Issue Plant Lipid Synthesis and Metabolism)
►▼
Show Figures

Figure 1
Open AccessArticle
A New Leaf Essential Oil from Endemic Gynoxys laurifolia (Kunth) Cass. of Southern Ecuador: Chemical and Enantioselective Analyses
Plants 2023, 12(15), 2878; https://doi.org/10.3390/plants12152878 - 06 Aug 2023
Abstract
The fresh leaves of Gynoxys laurifolia (Kunth) Cass. (Asteraceae), collected in the province of Loja (Ecuador), were submitted to steam distillation, producing an essential oil with a yield of 0.02% by weight. This volatile fraction, described here for the first time, was submitted to
[...] Read more.
The fresh leaves of Gynoxys laurifolia (Kunth) Cass. (Asteraceae), collected in the province of Loja (Ecuador), were submitted to steam distillation, producing an essential oil with a yield of 0.02% by weight. This volatile fraction, described here for the first time, was submitted to qualitative (GC–MS) and quantitative (GC–FID) chemical analyses, on two orthogonal columns (non-polar and polar stationary phase). A total of 90 components, corresponding to 95.9–95.0% by weight on the non-polar and polar stationary phase, respectively, were detected and quantified with at least one column. Major constituents (≥3%) were: germacrene D (18.9–18.0%), (E)-β-caryophyllene (13.2–15.0%), α-pinene (11.0–10.3%), β-pinene (4.5–4.4%), β-phellandrene (4.0–3.0%), bicyclogermacrene (4.0–3.0%), and bakkenolide A (3.2–3.4%). This essential oil was dominated by sesquiterpene hydrocarbons (about 45%), followed by monoterpene hydrocarbons (about 25–30%). This research was complemented with the enantioselective analysis of some common chiral terpenes, carried out through 2,3-diethyl-6-tert-butyldimethylsilyl-β-cyclodextrin and 2,3-diacetyl-6-tert-butyldimethylsilyl-β-cyclodextrin as stationary phase chiral selectors. As a result, (1S,5S)-(−)-β-pinene, (R)-(−)-α-phellandrene, (R)-(−)-β-phellandrene, (S)-(−)-limonene, (S)-(+)-linalyl acetate, and (S)-(−)-germacrene D were observed as enantiomerically pure compounds, whereas α-pinene, linalool, terpinene-4-ol, and α-terpineol were present as scalemic mixtures. Finally, sabinene was practically racemic. Due to plant wildness and the relatively low distillation yield, no industrial applications can be identified, in the first instance for this essential oil. The focus of the present study is therefore academic.
Full article
(This article belongs to the Collection Essential Oils of Plants (Chemical Composition, Variation and Properties))
►▼
Show Figures
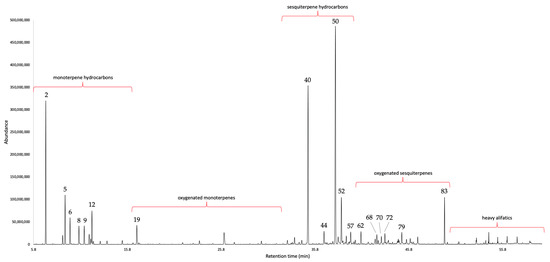
Figure 1

Journal Menu
► ▼ Journal Menu-
- Plants Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Diversity, Ecologies, Insects, Plants, Taxonomy
Biodiversity in the Azores: A Whole Biota Assessment
Topic Editors: Rosalina Gabriel, João Pedro Barreiros, Paulo A. V. BorgesDeadline: 15 August 2023
Topic in
Agriculture, Bioengineering, Genes, IJMS, Plants
Genetic Engineering in Agriculture
Topic Editors: Amy L. Klocko, Jianjun Chen, Haiwei LuDeadline: 31 August 2023
Topic in
Agronomy, Diversity, Ecologies, Microorganisms, Plants
Plant-Associated Microbiota: From the Assembly to the Function
Topic Editors: Alessandra Salvioli Di Fossalunga, Vincenza CozzolinoDeadline: 30 September 2023
Topic in
Agriculture, Agronomy, Crops, Plants, Viruses
Plant Virus
Topic Editors: Munir Mawassi, Sergey MorozovDeadline: 31 October 2023

Conferences
Special Issues
Special Issue in
Plants
Maintenance and Function of Biodiversity in Forests
Guest Editors: Xugao Wang, Zuoqiang Yuan, Chengjin ChuDeadline: 10 August 2023
Special Issue in
Plants
Plant Phytochemicals on Crop Protection and Drug Development
Guest Editors: Natália Cruz-Martins, Christophe HanoDeadline: 20 August 2023
Special Issue in
Plants
Wheat Crop Improvement (by Transgenic and Conventional Breeding Methods)
Guest Editors: Mark D. Wilkinson, Ondrej KosikDeadline: 31 August 2023
Special Issue in
Plants
Flax: A Traditional Culture with Modern Advantages
Guest Editors: Anthony Quéro, Christophe HanoDeadline: 20 September 2023
Topical Collections
Topical Collection in
Plants
Doubled Haploid Technology in Plant Breeding
Collection Editor: Thomas Lübberstedt
Topical Collection in
Plants
Feature Papers in Plant Physiology and Metabolism
Collection Editors: John T. Hancock, Mikihisa Umehara
Topical Collection in
Plants
Feature Papers in Plant Protection
Collection Editors: Paula Baptista, Livy Williams
Topical Collection in
Plants
Feature Papers in Plant Ecology
Collection Editors: Ismael Aranda, Fernando Henrique Reboredo, Roberta Masin